Monitoring Protocol
Purpose: to accurately and efficiently monitor spatial and temporal variation in fish abundance. The sampling design is focused primarily on detecting change in abundant to moderately abundant reef fish across all 24 study sites sampled on a bimonthly basis. The simple, low-replicate design minimizes site survey time while maximizing repeated surveying of sites to obtain estimates of temporal variation in adult and juvenile abundance.
A. Sampling Design
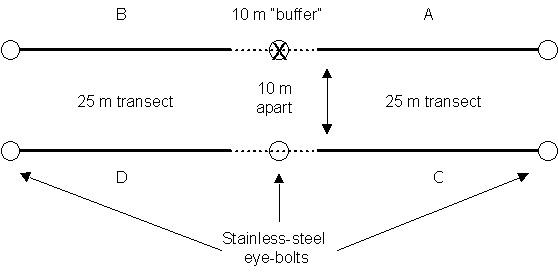
Design of transect layout used at permanent transects. Two 60 m neutral gray transect lines are positioned along six permanent stainless-steel eyebolts. Divers survey four 25 m transects separated by 10 m buffer areas between replicates.
B. Survey Protocol
Transects are located by differential GPS. Fish densities of all observed species are estimated by visual strip transect search along each permanent transect line. Two pairs of divers survey the lines, each pair searching two of the 25 m lines. The search of each line consists of two divers, swimming side-by-side on each side of the line, surveying a column 2 m wide. On the outward-bound leg, larger planktivores and wide-ranging fishes within 4 m of the bottom are recorded. On the return leg, fishes closely associated with the bottom, juveniles, and fishes hiding in cracks and crevices are recorded. Each site is scheduled to be surveyed bi-monthly, weather permitting, for a total of six surveys per year.
C. Power Analysis (based on first survey)
| Mean Density = | per 100m2 |
| MDD = |
Minimum Detectable Difference of design (in fish per 100m2) with a=0.05 b=0.10. |
| % Change = | detectable percent change in abundance. Change greater than this between surveys would result in a statistically significant difference. |
| Species | Mean Density |
Standard Deviation |
MDD | % Change |
| Aquarium Species | ||||
|
Acanthurus achilles |
0.86 |
1.03 |
0.41 |
47 |
|
Acanthurus olivaceus |
0.85 |
1.81 |
0.72 |
85 |
|
Centropyge potteri |
2.09 |
1.30 |
0.52 |
25 |
|
Chaetodon auriga |
0.09 |
0.29 |
0.11 |
132 |
|
Chaetodon fremblii |
0.04 |
0.21 |
0.08 |
191 |
|
Chaetodon lunula |
0.33 |
0.58 |
0.23 |
70 |
|
Chaetodon multicinctus |
5.78 |
1.67 |
0.66 |
11 |
|
Chaetodon ornatissimus |
1.66 |
1.24 |
0.49 |
30 |
|
Chaetodon quadrimaculatus |
0.80 |
1.02 |
0.41 |
51 |
|
Chaetodon unimaculatus |
0.09 |
0.29 |
0.11 |
132 |
|
Ctenochaetus hawaiiensis |
0.65 |
0.97 |
0.39 |
59 |
|
Ctenochaetus strigosus |
21.67 |
9.03 |
3.60 |
17 |
|
Zebrasoma flavescens |
14.46 |
9.66 |
3.85 |
27 |
| Non-Aquarium Species | ||||
|
Abudefduf abdominalis |
1.00 |
2.10 |
0.84 |
84 |
|
Acanthurus blochii |
0.04 |
0.21 |
0.08 |
191 |
|
Acanthurus leucopareius |
0.17 |
0.65 |
0.26 |
149 |
|
Acanthurus nigricans |
0.30 |
0.63 |
0.25 |
83 |
|
Acanthurus nigrofuscus |
9.03 |
4.74 |
1.89 |
21 |
|
Acanthurus nigroris |
1.27 |
1.35 |
0.54 |
42 |
|
Acanthurus thompsoni |
2.08 |
3.40 |
1.35 |
65 |
|
Acanthurus triostegus |
0.61 |
1.77 |
0.71 |
116 |
|
Aluterus scriptus |
0.04 |
0.21 |
0.08 |
191 |
|
Anampses cuvier |
0.04 |
0.21 |
0.08 |
191 |
|
Aphareus furca |
0.09 |
0.29 |
0.11 |
132 |
|
Apogon kallopterus |
0.13 |
0.63 |
0.25 |
191 |
|
Apogon menesemus |
0.13 |
0.46 |
0.18 |
140 |
|
Apogon spp. |
0.22 |
0.60 |
0.24 |
110 |
|
Arothron meleagris |
0.13 |
0.34 |
0.14 |
105 |
|
Aulostomus chinensis |
0.40 |
0.60 |
0.24 |
59 |
|
Balistes polylepis |
0.04 |
0.21 |
0.08 |
191 |
|
Bodianus bilunulatus |
0.13 |
0.34 |
0.14 |
105 |
|
Calotomus carolinus |
0.09 |
0.29 |
0.11 |
132 |
|
Cantherhines dumerilii |
0.35 |
0.71 |
0.28 |
82 |
|
Canthigaster amboinensis |
0.13 |
0.46 |
0.18 |
140 |
|
Canthigaster jactator |
2.51 |
2.87 |
1.14 |
46 |
|
Caracanthus typicus |
0.04 |
0.21 |
0.08 |
191 |
|
Cephalopholis argus |
0.93 |
0.65 |
0.26 |
28 |
|
Chaetodon lineolatus |
0.04 |
0.21 |
0.08 |
191 |
|
Chaetodon miliaris |
0.32 |
1.17 |
0.47 |
146 |
|
Chromis agilis |
20.50 |
16.54 |
6.59 |
32 |
|
Chromis hanui |
3.36 |
2.30 |
0.92 |
27 |
|
Chromis ovalis |
10.46 |
36.16 |
14.40 |
138 |
|
Chromis vanderbilti |
20.06 |
31.66 |
12.61 |
63 |
|
Chromis verater |
1.01 |
2.34 |
0.93 |
92 |
|
Cirrhitops fasciatus |
0.20 |
0.54 |
0.21 |
110 |
|
Cirrhitus pinnulatus |
0.17 |
0.49 |
0.20 |
112 |
|
Cirripectes vanderbilti |
0.17 |
0.39 |
0.15 |
89 |
|
Coris flavovittata |
0.17 |
0.39 |
0.15 |
89 |
|
Coris gaimard |
0.87 |
0.77 |
0.31 |
35 |
|
Coris venusta |
0.13 |
0.34 |
0.14 |
105 |
|
Dascyllus albisella |
0.91 |
1.62 |
0.65 |
71 |
|
Exallias brevis |
0.37 |
0.61 |
0.24 |
65 |
|
Fistularia commersonii |
0.17 |
0.49 |
0.20 |
112 |
|
Forcipiger flavissimus |
0.92 |
1.07 |
0.42 |
46 |
|
Forcipiger longirostris |
0.96 |
1.12 |
0.44 |
47 |
|
Gomphosus varius |
1.80 |
1.41 |
0.56 |
31 |
|
Gymnothorax flavimarginatus |
0.17 |
0.39 |
0.15 |
89 |
|
Gymnothorax meleagris |
0.30 |
0.56 |
0.22 |
73 |
|
Gymnothorax spp. |
0.09 |
0.42 |
0.17 |
191 |
|
Halichoeres ornatissimus |
1.84 |
1.24 |
0.50 |
27 |
|
Hemitaurichthys polylepis |
0.78 |
2.32 |
0.92 |
118 |
|
Hemitaurichthys thompsoni |
0.61 |
2.31 |
0.92 |
151 |
|
Labroides phthirophagus |
1.87 |
0.96 |
0.38 |
20 |
|
Lutjanus kasmira |
0.22 |
0.74 |
0.29 |
135 |
|
Macropharyngodon geoffroyi |
0.13 |
0.34 |
0.14 |
105 |
|
Melichthys niger |
2.06 |
3.51 |
1.40 |
68 |
|
Melichthys vidua |
0.71 |
0.71 |
0.28 |
40 |
|
Monotaxis grandoculis |
0.49 |
1.28 |
0.51 |
105 |
|
Mulloidichthys flavolineatus |
0.09 |
0.42 |
0.17 |
191 |
|
Mulloidichthys vanicolensis |
5.61 |
14.04 |
5.59 |
100 |
|
Myripristis amaena |
0.13 |
0.63 |
0.25 |
191 |
|
Myripristis berndti |
4.12 |
7.15 |
2.85 |
69 |
|
Myripristis kuntee |
2.01 |
4.24 |
1.69 |
84 |
|
Naso hexacanthus |
0.22 |
0.52 |
0.21 |
95 |
|
Naso lituratus |
1.99 |
1.54 |
0.61 |
31 |
|
Neoniphon sammara |
0.43 |
0.90 |
0.36 |
82 |
|
Novaculichthys taeniourus |
0.04 |
0.21 |
0.08 |
191 |
|
Ostracion meleagris |
0.22 |
0.50 |
0.20 |
91 |
|
Oxycheilinus unifasciatus |
0.67 |
0.60 |
0.24 |
35 |
|
Paracirrhites arcatus |
5.69 |
6.76 |
2.69 |
47 |
|
Paracirrhites forsteri |
1.04 |
0.73 |
0.29 |
28 |
|
Parupeneus bifasciatus |
0.91 |
1.12 |
0.45 |
49 |
|
Parupeneus cyclostomus |
0.26 |
0.75 |
0.30 |
115 |
|
Parupeneus multifasciatus |
1.26 |
0.99 |
0.39 |
31 |
|
Parupeneus porphyreus |
0.04 |
0.21 |
0.08 |
191 |
|
Pervagor aspricaudus |
0.51 |
1.02 |
0.41 |
80 |
|
Plagiotremus goslinei |
0.26 |
0.54 |
0.22 |
83 |
|
Plectroglyphidodon imparipennis |
0.22 |
0.52 |
0.21 |
95 |
|
Plectroglyphidodon johnstonianus |
2.57 |
1.22 |
0.49 |
19 |
|
Priacanthus meeki |
0.04 |
0.21 |
0.08 |
191 |
|
Pseudocheilinus evanidus |
2.19 |
1.29 |
0.51 |
23 |
|
Pseudocheilinus octotaenia |
3.29 |
1.79 |
0.71 |
22 |
|
Pseudocheilinus tetrataenia |
1.91 |
1.32 |
0.53 |
27 |
|
Pseudojuloides cerasinus |
0.26 |
0.75 |
0.30 |
115 |
|
Sargocentron ensiferum |
0.26 |
0.92 |
0.36 |
140 |
|
Sargocentron spiniferum |
0.09 |
0.29 |
0.11 |
132 |
|
Sargocentron spp. |
0.22 |
0.67 |
0.27 |
123 |
|
Sargocentron tiere |
0.04 |
0.21 |
0.08 |
191 |
|
Sargocentron xantherythrum |
0.26 |
0.69 |
0.27 |
105 |
|
Saurida gracilis |
0.04 |
0.21 |
0.08 |
191 |
|
Scarus dubius |
0.13 |
0.46 |
0.18 |
140 |
|
Scarus perspicillatus |
0.04 |
0.21 |
0.08 |
191 |
|
Scarus psittacus |
0.20 |
0.94 |
0.37 |
191 |
|
Scarus rubroviolaceus |
0.48 |
1.08 |
0.43 |
90 |
|
Scarus sordidus |
2.50 |
3.70 |
1.47 |
59 |
|
Scorpaenopsis cacopsis |
0.09 |
0.29 |
0.11 |
132 |
|
Scorpaenopsis diabolus |
0.04 |
0.21 |
0.08 |
191 |
|
Stegastes fasciolatus |
1.86 |
2.77 |
1.10 |
59 |
|
Stethojulis balteata |
0.49 |
0.61 |
0.24 |
49 |
|
Sufflamen bursa |
1.54 |
0.83 |
0.33 |
21 |
|
Synodus dermatogenys |
0.04 |
0.21 |
0.08 |
191 |
|
Synodus spp. |
0.04 |
0.21 |
0.08 |
191 |
|
Synodus ulae |
0.04 |
0.21 |
0.08 |
191 |
|
Synodus variegatus |
0.04 |
0.21 |
0.08 |
191 |
|
Thalassoma ballieui |
0.04 |
0.21 |
0.08 |
191 |
|
Thalassoma duperrey |
6.51 |
2.58 |
1.03 |
16 |
|
Xanthichthys auromarginatus |
0.73 |
1.35 |
0.54 |
74 |
|
Zanclus cornutus |
0.73 |
0.87 |
0.35 |
47 |
D. Coral Assessment Protocol
We will utilize standard methods developed in conjunction with the Hawai'i Coral Reef Assessment and Monitoring Program (CRAMP) to estimate substratum abundance, diversity and distribution. Currently these include digital video imaging to record 50 contiguous frames along each transect line (n=200 photographs for each site). Images are analyzed to obtain percent cover estimates for substratum types. See coral monitoring for more information.
